
(C) 2002 G. F. Schröder and H. Grubmüller (Göttingen)
Overview
 |

(C) 2002 G. F. Schröder and H. Grubmüller (Göttingen) |
|
Overview
|
|
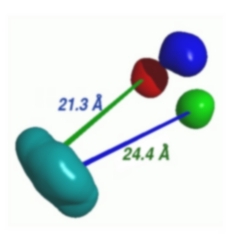
| FRETsg is a tool for a fast qualitative analysis of multiple FRET (fluorescence resonance energy transfer) experiments. FRET experiments principally yield the distances between fluorescent dyes, which are commonly used for labelling proteins, DNA, RNA, etc. If multiple FRET experiments are done, it is possible to build a 3D model of the labelled positions. FRETsg finds all positions which obey the measured FRET distances and writes the model to a PDB-type file, which can be easily overlayed with other PDB structures. | |
| Download | |
| Manual | |
| License | |
FRET structure generator, distance,
dyes, fluorophores, protein structure determination, protein fluctuation
analysis, triangulation, Gunnar Schröder, Gunnar Schroder, Helmut
Grubmuller,
Helmut Grubmüller. fluorescence
resonance energy transfer experiments. forster transfer, förster transfer,
förster theory, forster theory,